Tree alignment
In computational phylogenetics, tree alignment is the problem of producing a multiple sequence alignment on a set of sequences over a fixed tree.
Formally, tree alignment is the following optimization problem.
Input: A set 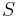 of sequences, a phylogenetic tree
of sequences, a phylogenetic tree 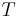 leaf-labeled by
leaf-labeled by 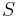 and an edit distance function
and an edit distance function 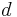 between sequences,
between sequences,
Output: A labeling of the internal vertices of 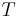 such that
such that 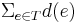 is minimized, where
is minimized, where 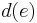 is the edit distance between the endpoints of
is the edit distance between the endpoints of 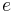 .
.
See also
References
- ^ Elias, Isaac (2006), "Settling the intractability of multiple alignment", J Comput Biol 13 (7): 1323–1339, doi:10.1089/cmb.2006.13.1323, PMID 17037961